In late December 2019, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan, China, with human respiratory problems resulting from the likely transmission of an animal-adapted coronavirus.
This was the first time in decades that pandemics have resurfaced as a global problem. There were, however, early warnings of a global event with SARS and avian flu. Early containment in both cases was successful, but the strategy for SARS-CoV-2 shifted to mitigation after early containment failed. Almost all countries observed this pattern at the beginning of the introduction of COVID-19 across national borders.
As a result of human-animal transmission, there are grave concerns regarding the possibility of a re-ignition of the COVID pandemic through resistance mutations cultivated in animal reservoirs.
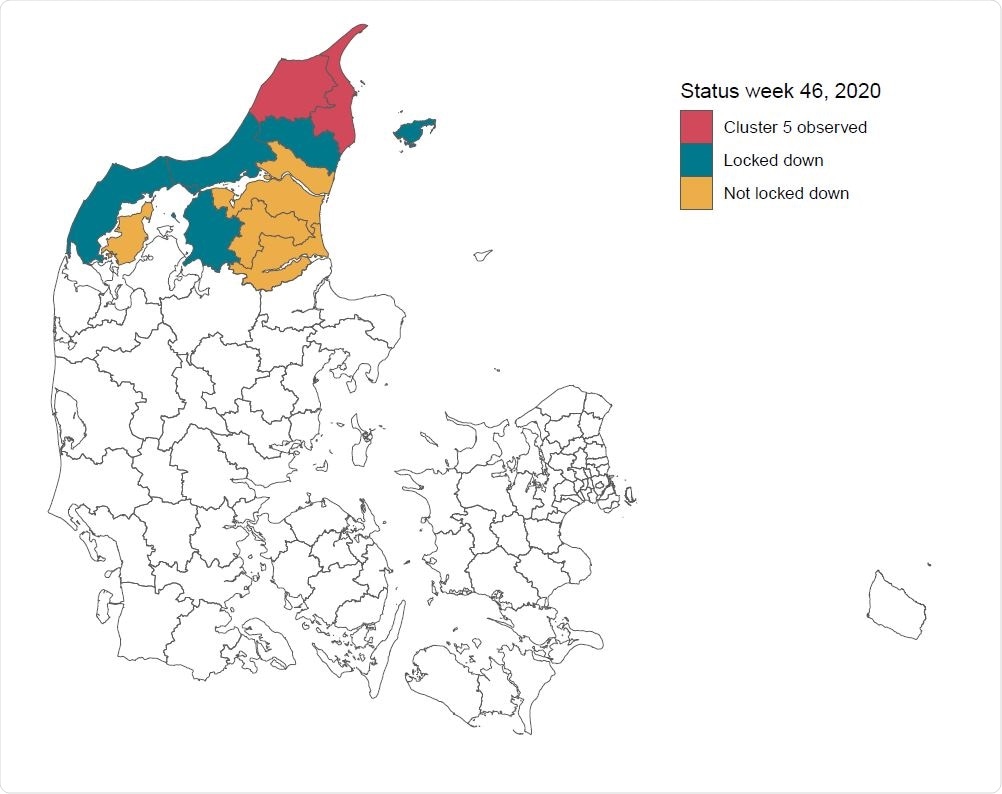
One specific example of animal-human transmission being the cluster-5 mutation that was found to have transferred from minks farmed in the Danish fur industry in mid-2020. The cluster-5 variant of SARS-CoV-2 was thought to have moderately reduced sensitivity to antibodies, and thus decisive action was taken with regards to strict containment measures, lasting for four weeks, after which the variant has not been observed again in mink or humans.
However, it cannot be confirmed for certain that the strain has been eliminated, raising the question of how long a strain must go undetected before being considered, in all probability, extinct.
In a paper recently uploaded to the preprint server medRxiv* by Schiøler et al. (July 6th, 2021), mathematical models are employed to probe this question, using the case study of the cluster-5 variant to test them.
.jpg)
How was the model assembled?
Several epidemiology modeling techniques are available, some more suitable to modeling populations of larger or smaller size and complexity. The group elected to use a Markov model, which incorporates pseudo-randomly changing parameters in the modeling system, one example being the SARS-CoV-2 reproduction number.
The calculation for the probability of lineage extinction over time formulated by the group considers the true and observed number of infected people carrying the variant, birth, and death rate of the variant, and population size at a particular time.
In all simulations, the recovery time from SARS-CoV-2 was set to two weeks, the usual period of infectivity for the virus, and as a test case, the numbers involved in the cluster-5 outbreak were entered.
The total population considered was 600,000, reflecting the northern Denmark region, with 10,000 SARS-CoV-2 PCR tests being performed per week, along with a rising number of whole-genome sequencing that reflected the numbers recorded by the Danish government. The initial number of infected was set to 11, those that were detected with the lineage at the initial stages of the outbreak.
The initial lockdown was planned to last for four weeks, though the strictest measures were dropped after two, following political pressure. The variant was not detected again in the population, however, suggesting it had been contained. According to the formula developed by the research group, the probability that the lineage was already extinct upon beginning non-pharmaceutical interventions was 0.22, rising to 0.37 two weeks later when restrictions were lifted. If the lockdown had been lifted when it was originally intended, four weeks later, the probability that the lineage would be extinct rose to 0.7.
Further implications
Applying the model to the known Danish case provided some context to the methods developed, and the group notes other interesting trends associated with the probability of lineage extinction.
The effect of increasing the whole genome testing rate amongst the population while keeping the reproduction number of the virus constant was investigated, finding that the number of weeks until extinction lowers from 42 to 25 by increasing the testing rate. This is likely due to the greater testing rate identifying individuals carrying the specific strain instead of PCR testing only that could not distinguish between strains and allow the tested individuals to be isolated quickly.
They also probed the probability of extinction when increasing reproduction rates while keeping the testing rate constant, noting an interesting relationship wherein the maximum number of weeks to extinction was greatest at reproduction rates slightly less than 1. That is, at higher SARS-CoV-2 reproduction rates the number of weeks it would take until the extinction of the specific SARS-CoV-2 lineage while keeping testing constant, is shorter than at rates of approximately 0.9.
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Mathematical modelling of SARS-CoV-2 variant outbreaks reveals their probability of extinction, Henrik Schiøler, Torben Knudsen, Rasmus Froberg Brøndum, Jakob Stoustrup, Martin Bøgsted, medRxiv, 2021.07.05.21260005; doi: https://doi.org/10.1101/2021.07.05.21260005, https://www.medrxiv.org/content/10.1101/2021.07.05.21260005v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Antibodies, Coronavirus, Coronavirus Disease COVID-19, Epidemiology, Flu, Genome, Mutation, Pandemic, Reproduction, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Virus

Written by
Michael Greenwood
Michael graduated from Manchester Metropolitan University with a B.Sc. in Chemistry in 2014, where he majored in organic, inorganic, physical and analytical chemistry. He is currently completing a Ph.D. on the design and production of gold nanoparticles able to act as multimodal anticancer agents, being both drug delivery platforms and radiation dose enhancers.
Source: Read Full Article
